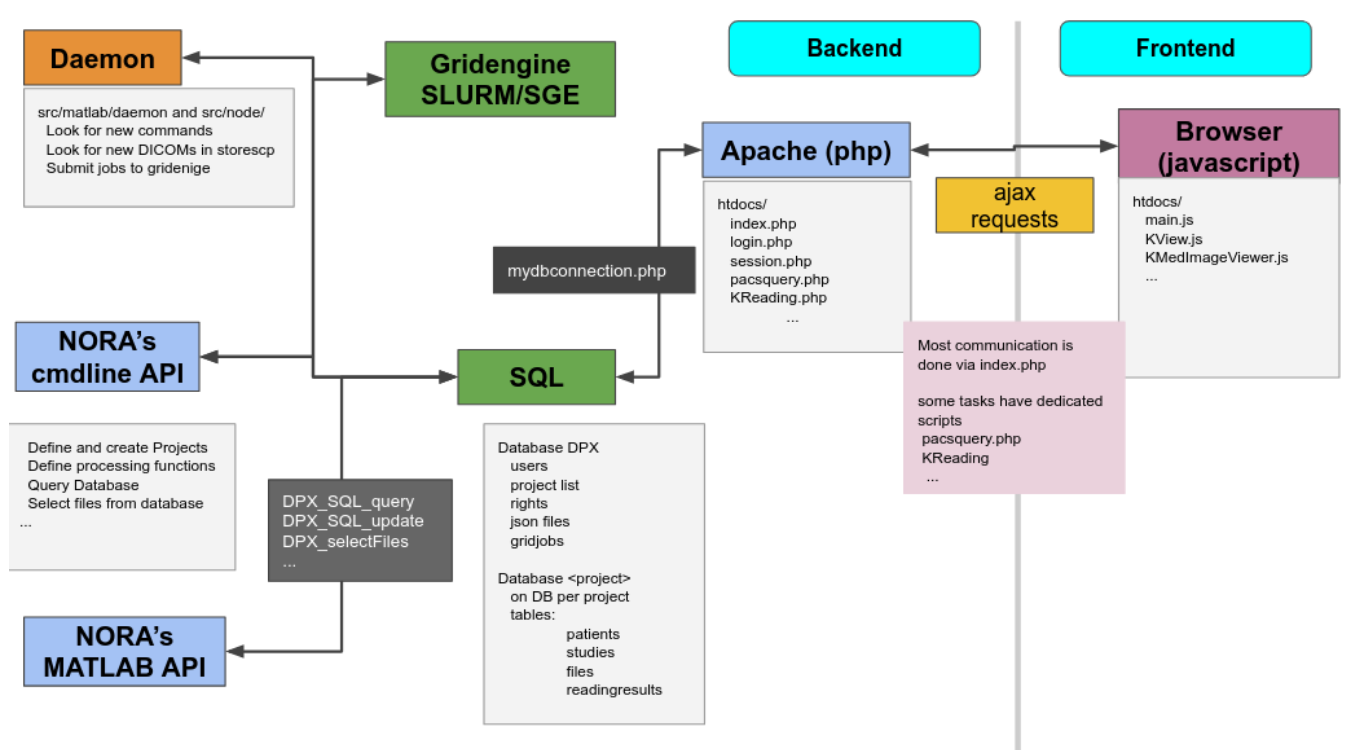
Administration Backend
Command line Interface
You need a PATH to <path-to-nora>/src/node
Call nora
And some more intro to project jsons …..
NORA - backend
usage: nora [--parameter] [value]
--project (-p) [name]
all preceding commands refer to this project
MANAGEMENT
--sql (-q) [sqlstatement] [--csv]
executes SQL statment and return result as json (or as csv if option is given)
--add (-a) [file] ...
adds list of files to project
--del (-d) [file] ...
deletes list files
--out (-o) [filepattern]
computes absolute outputfilepath from filepattern (see description of filepattern below)
--pathout (-pa) [filepattern]
computes absolute outfilepath from filepattern
--select (-s) [--json (-j)] [filepattern]
returns files matching filepattern, if --json is given, files are return as json filepattern
--launch (-l) [json]
launches batch from json description
--createproject (-c) [name] [module]
creates a project with [name] from [module] definition
--admin [parameter1] [parameter2] ...
calls the admin tools (see nora --admin for more help)
PROCESSING
--launch json
launches a NORA job given by json
options: --throwerror
--handle_job_result (emits DONE signal, when ready)
--launchfile jsonfile
same as --launch but json given by fileref
--launch_autoexec psid
launches the autoexecution pipeline of the project for the patient specified by psid
--import srcpath
imports dicoms located in sourcepath to given project
options: --autoexec_queue (if given autoexecuter is initiated, use DEFAULT to select default autoexecution queue given in config)
--handle_job_result (emits DONE signal, when ready)
--extrafiles_filter: if there are non-dicom files in your dicomfolder and you
want to add them to your patient folder, then, give here a comma-separated list of
accepted extensions (e.g. nii,hdr,img)
--patients_id PIZ (overwrite patients_id of imported data with PIZ )
--studies_id SID (overwrite studies_id of imported data with SID )
--pattern REGEX (use REGEX applied on srcpath of dicoms to overwrite
patients_id/studies_id, example:
--pattern "(?<studies_id>[\w\-]+)\/(?<patients_id>\w+)$"
OTHER
--exportmeta [filepattern]
exports meta data as csv. Meta content from files matching [filepattern] are exported
options: --keys a comma separated list of key-sequences, which are exported [optional]
a key sequence is given by a dot-separated list, where the first key refers to the name of
the json-file (as multiple jsons might be contained in the query). A wildcard * is possible
select multiple keys at once
examples:
nora -p XYZ --exportmeta '*' 'nodeinfo.json' // gathers also patient info
nora -p XYZ --exportmeta '*' 'META/radio*.json' --keys '*.mask.volume,*.mask.diameter'