The Batchtool
The Batchtool Window
Consider Figure 2 below: the subject/studies table on the left is used for the selection of subject/studies on which you want to run your batch. You can use the filter bars to create the subgroup you want to work on (see Subject/Studies table). Select "Batchtool" on the top toolbar (A) to open the batchtool. Figure 2 shows the structure of the Bacthtool window. It allows to compose the batch out of single jobs. Jobs are added from the menu (D). You can save batches (E), which then appear in the batch list (C). To open an overview of currently running jobs open the "Gridstats" window (B) or (H).
Batches are launched for every subjects/study independently in parallel. The jobs within a batch run sequentially. You can also choose different running option (see (G) in Figure 2). Depending on the selection level (subjects or studies), the batches are iterated over subjects or patients
![]()
Imagine a scenario where you have multiple studies per patient, which have to be linked in some sense. Then, the subject level is appropriate. For example, think of a neuroimaging analysis where you a have a CT study (which contains, e.g. electrode information) and a MR study (which contains soft tissue anatomical information), or think of a simple longitudinal analysis. Otherwise, if your your studies should all be treated in an equal manner, the study level is appropriate.
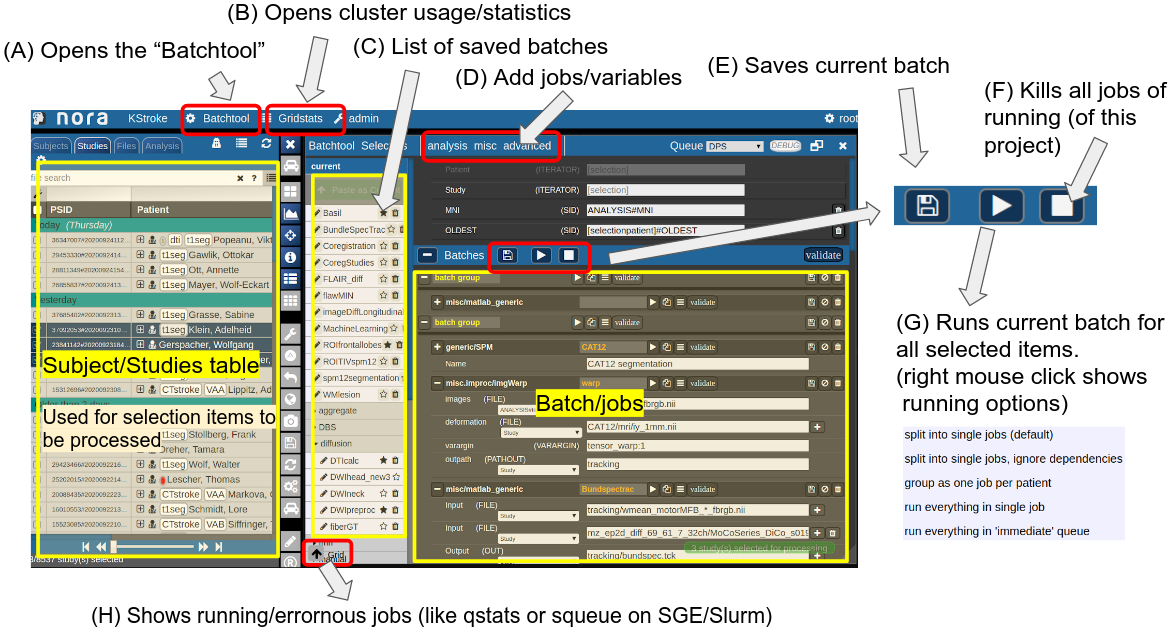 Figure 2: Batchtool overview.
Figure 2: Batchtool overview.
The Anatomy of a Job
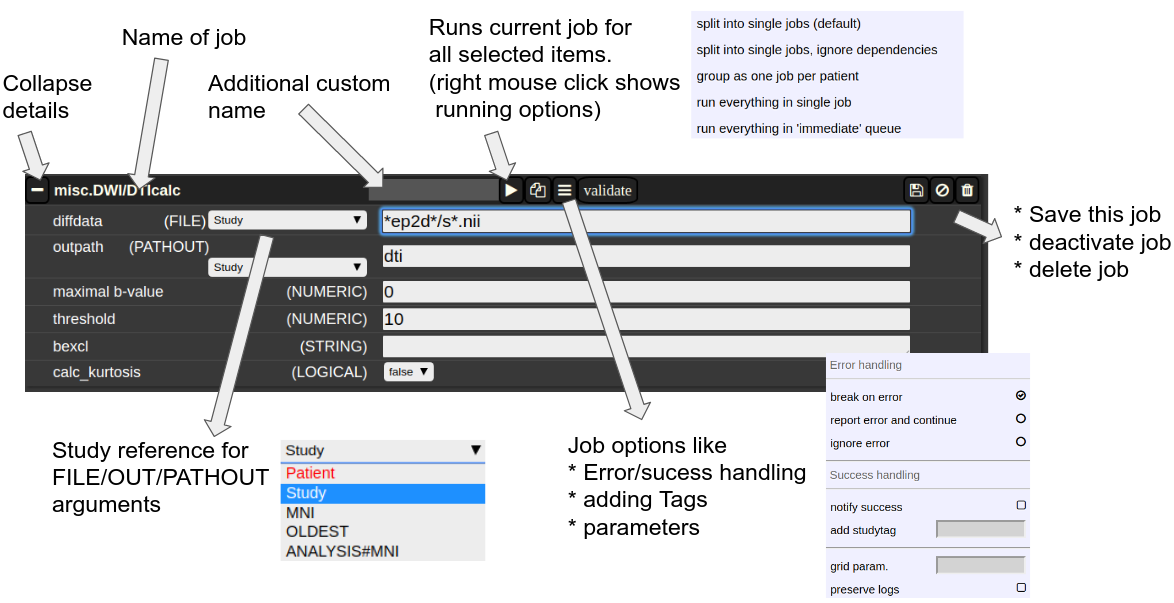
A job consists of a list of arguments. There are several types of arguments:
-
FILE
All input images/series (or any other type of files) are given as FILE arguments. Usually you give a file pattern instead of an explicit filename. A fIle pattern is a combination of subfolders, filename and wildcards. For example: t1*/s0__.nii. I refers to all files contained in a folder starting with t1 and whose filename matches "s0___.nii" . The asterisks (*) is a placeholder for an arbitrary character sequence, an underscore "_" for a single character. Internally, the wildcards are the same as for SQL "like" statement (the '*' is replaced by '%'). A FILE argument also includes a reference to a study or patient. Depending on the selection level (subject or study), different "study references" are possible. See below for more about "study references". -
OUT
A name of a file including the subfolder. No wildcards are allowed here. Depending on the selection level there are again different study references possible. -
PATHOUT
Same as OUT but refers to foldername instead of a filename. - NUMERIC
- STRING
- LOGICAL
- OPTION
Study References and study selectors
Figure 3: The anatomy of a single job.
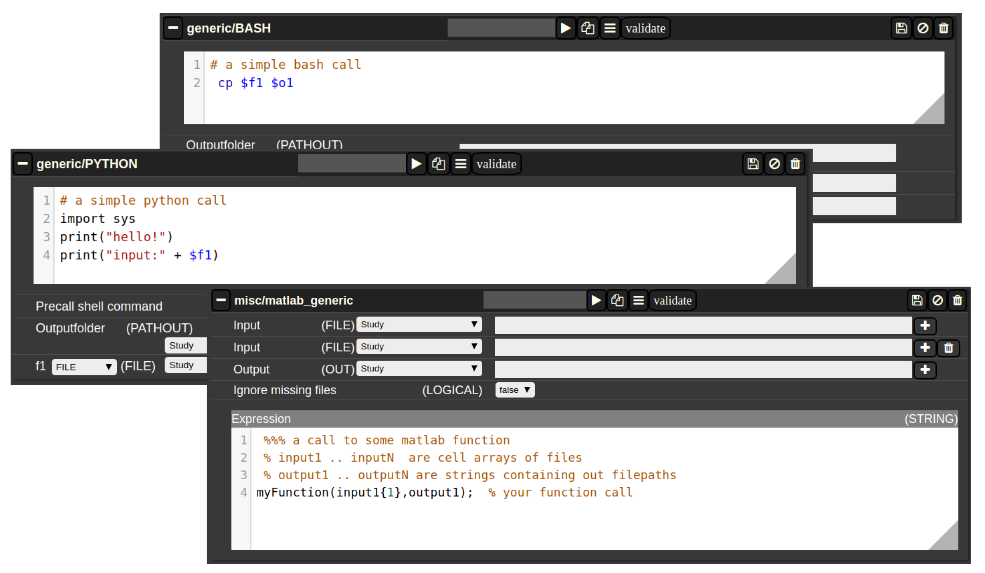
Generic jobs
There are a multitude of predefined algorithms (mostly MATLAB) in NORA; however you can also implement your own scripts directly by using generic jobs. Currently there are three types of languages possible:
- BASH
- Python
- MATLAB
A generic jobs basically provides a field where you can enter simple expression or a full script in BASH/Python or MATLAB. Arguments from NORA are passed to the script by simple variable naming conventions.
For BASH/Python scripts input files (and all other parameters) are referenced by variables with a $-prefix with a special naming convention. For example, file arguments are referenced by $f1-$f9. Once NORA finds such an expression it automatically adds a corresponding row at the bottom of the job, which can be filled by the appropriate file patterns. The same holds of output arguments (represented by $o1-$o9) and output paths (prefix 'p'). Other parameters (STRING,NUMERIC) are referenced by prefixes 's' and 'n'.
In MATLAB the approach is a little bit different. You can manually add input/output arguments by using the "plus" sign and refer to the arguments by ordinary MATLAB variables. As input you have a series of cell-Arrays (input1, ..., inputN), as output a series of strings (output1, ..., outputN).
Figure 4: Generic Jobs
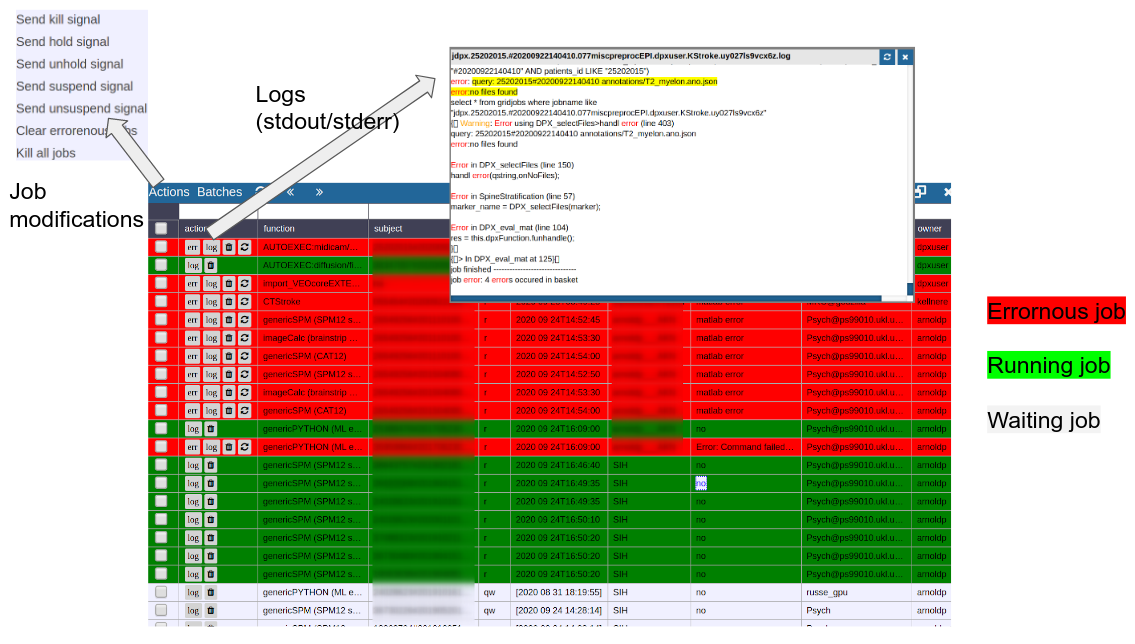
Cluster Managment/Monitor (Gridstats)
To monitor the integrated cluster environmenr there is a simple table based overview, which provides access job logs and job modifications. One can also sort and search the current job load to selectively monitor or kill/suspend jobs. While finished jobs just disappear by default (you can change this in the settings), jobs that have produced an error are kept for further analysis. Note that the job information is also available on subject/study level (see Figure 2) as small indicators. To get further information about the job, you can click on the function cell and a JSON-representation of the job is displayed. A click on the subject cell selects the corresponding subject/study in the table.
Figure 5: Gridstats: monitoring and control of the cluster environment/resource managment (Slurm/SGE)